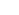
Developing Organ-scale Histology Techniques
Visualizing and labeling fixed biological organs in 3D allows us to assess 3D landscape of structural and molecular properties of every cells in organs. It is especially beneficial to study the brain since the brain is composed of neural network whose structure is in 3D. To facilitate this goal, we develop Organ-scale histology techniques
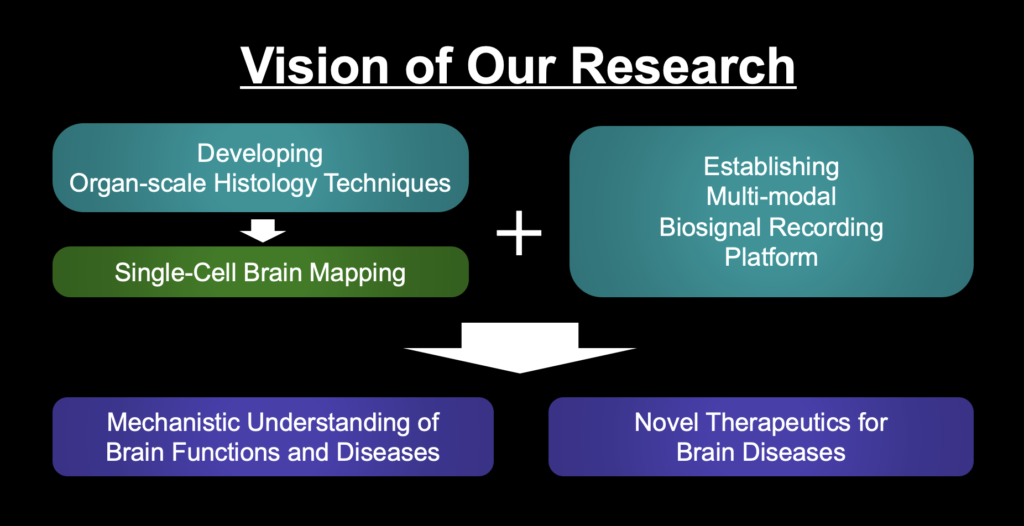
SHIELD
SHIELD is an Organ-scale tissue visualization technique (Park et al., 2019, Nature Biotechnology). It protects biological organs using special crosslinking chemistry to preserve biomolecules and tissue structure while making tissue transparent. It also can expand tissue for 3D super-resolution imaging of tissues (Fig. 1).
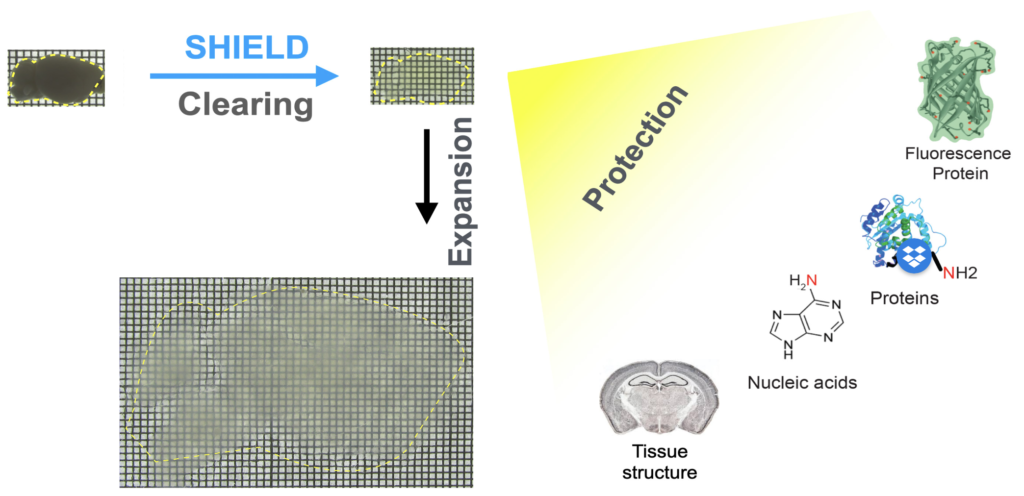
Fig. 1. SHIELD
eFLASH
eFLASH is an organ-scale immunolabeling technique that enables rapid, cost-effective, and universal 3D immunolabeling (Yun et al., 2019, Biorxiv). eFLASH achieves this by using rapid antibody transport with electrophoresis and the dynamic modulation of antibody binding (Fig. 2, left). eFLASH is compatible with ~150 antibodies, and diverse organs/tissues (Fig. 2, right).
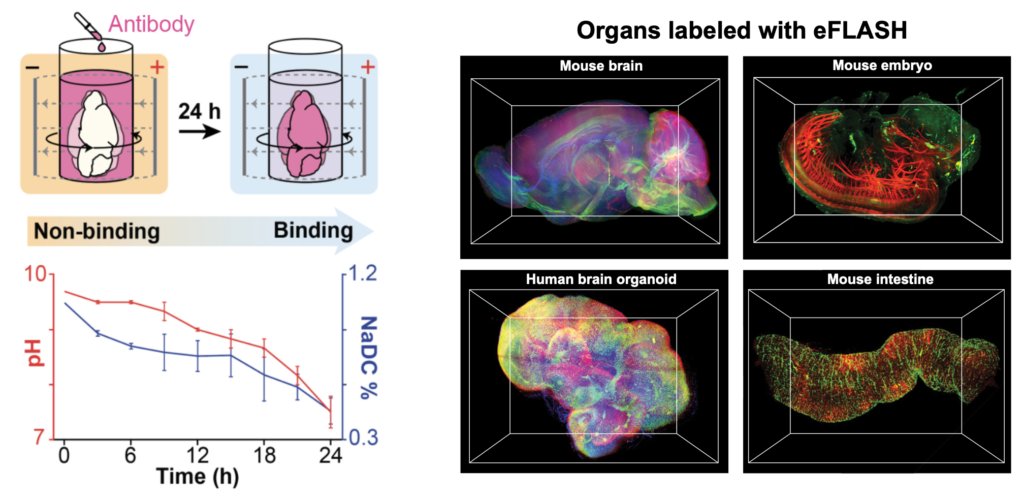
Fig. 2. eFLASH
Single-cell Brain Mapping
SHIELD and eFLASH are great tools for realizing single-cell brain mapping, the mapping of the brain with subcellular resolution. Two techniques can be connected to compose a pipeline(Fig. 3). A brain from an alive animal can be protected, cleared, and expanded using SHIELD, labeled using eFLASH, and then be imaged using high-speed fluorescence microscopy. Whole brain images can be analyzed using AI tools such as machine learning.
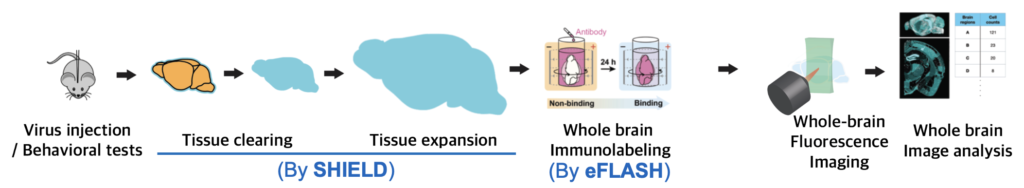
Fig. 3 Our Single-cell brain mapping pipeline
Below is an example of single-cell brain mapping in our group (Video 1).
Video 1. Counting PV+ cells from individual brain regions using machine learning-based algorithm.
We also use our pipeline for mapping neural network. Below is an example (Fig. 4.).
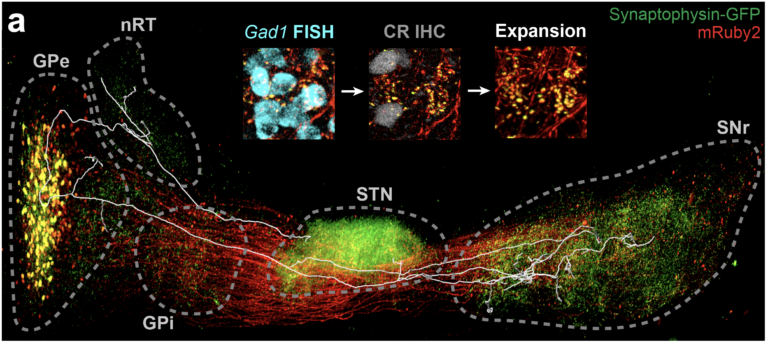
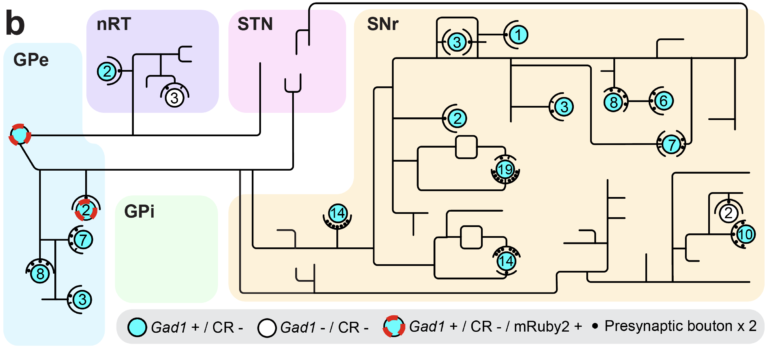
Fig. 4. Neural network mapping. (a) Labeled neural network with a trace of single neuron and (b) its map.
Establishing Multi-modal Biosignal Recording Platform
Our Single cell brain mapping extracts structural and molecular information from fixed(‘ex vivo’) brain. To understand brain functions and dysfunctions, functional information needs to be further extracted. Thus, we are establishing a platform(Fig. 5) that can extract multi-modal biosignals from alive mice (‘in vivo‘), and aim to integrate structural, molecular, and functional information altogether.
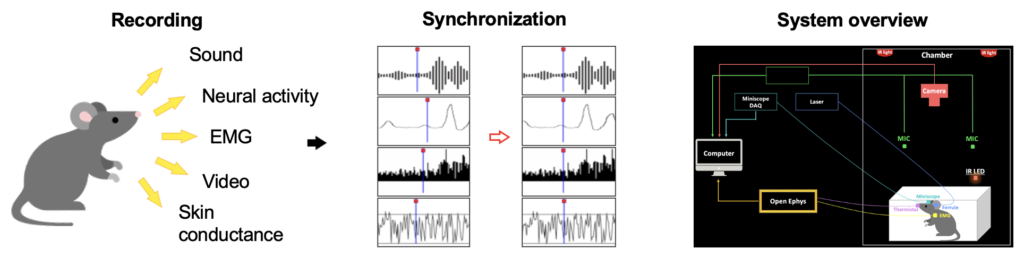
Fig. 5. Our Multi-modal Biosignal Recording Platform
Mechanistic Understanding of Brain Functions
Combining Our Organ-scale histology techniques and Multi-modal biosignal recording platform, we aim to mechanistically understand brain functions and diseases, focusing on memory and motor functions, and Depression and Alzheimer’s disease, respectively. As an example, we recently revealed that neurons storing a specific memory (“engram cells”) are distributed throughout the brain (Video 2; Roy et al., 2022, Nature Communication)
Video 2. Mapped engram cells of contextual fear memory
Novel Therapeutics for Brain diseases
By mapping novel brain networks and engineering their processes, we aim to devise safe and specific therapeutics for brain diseases.
*For detailed explanation of our SHIELD and eFLASH technques, please refer to this news article
https://www.ksmcb.or.kr/webzine/2106/content/research_01.html
- 1. Park Y-G*, Sohn CH*, Chen R*, McCue M, Yun DH, Drummond GT, Ku T, Evans NB, Oak HC, Trieu W, Choi H, Jin X, Lilascharoen V, Wang J, Truttmann MC, Qi HW, Ploegh HL, Golub TR, Chen S-C, Frosch MP, Kulik HJ, Lim BK, Chung K (2019) Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nature Biotechnology, 37(1), pp 73–83 (Cover article)
- 2. Yun DH*, Park Y-G*, Cho JH*, Kamensky L, Evans NB, Albanese A, Xie K, Swaney J, Sohn CH, Tian Y, Zhang Q, Drummond G, Guan W, DiNapoli N, Choi H, Jung H-Y, Ruelas L, Feng G, Chung K (2019) Ultrafast immunostaining of organ-scale tissues for scalable proteomic phenotyping. Under revision in Nature Biotechnology; bioRxiv, 660373.
- 3. Roy DS*, Park Y-G*, Ogawa SK*, Cho JH, Choi H, Kamensky L, Martin J, Chung K, Tonegawa S (2019) Brain-wide mapping of contextual fear memory engram ensembles supports the dispersed engram complex hypothesis. bioRxiv, 668483.
- 4. Park Y-G, Choi JH, Lee C, Kim S, Kim Y, Chang K-Y, Paek SH, Kim D. (2015) Heterogeneity of tremor mechanisms assessed by tremor-related cortical potential in mice. Molecular Brain, 8(1), p 3