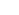Bio-Information System Laboratory (BISL), designated as a national research laboratory in 2005, has been developing innovative bioinformatics technologies and applying the technologies to medical and scientific applications. It has developed MONET to infer large-scale genetic networks from gene expression profiles by using Bayesian networks. The software is currently distributed through the international Cytoscape consortium. The laboratory has conducted international collaboration with Univ. of California at San Diego and Samsung Medical Center, and introduced a concept of network markers, which can resolve the shortcomings of conventional gene markers for disease diagnosis. The technology has been cited around 2,000 times according to the Google citation analysis. BISL is also leading a national research project called ‘Bio-Synergy Research Project’, where multi-component-multi-target effects of drugs and functional foods are analyzed with computerized virtual human systems called CODA along with structured assays of cell-based, animal-based, and clinical experiments. It is actively developing big bio-data mining and systems bioinformatics technologies to uncover hidden but essential rules behind various bio-medical phenomena. BISL is fostering passionate leaders creating new industries in the bio-medical fields
Network Bioinformatics Sponsors: Bio-Synergy Research Project of the Ministry of Science and ICT
Network Bioinformatics team is developing virtual physiological human systems that can be utilized in interpreting traditional medicine or natural compound efficacy and analyzing activation mechanisms based on computational approaches. To achieve these goals, we collect and integrate various types of biological data set encompassing protein-protein interaction networks, metabolic networks, gene regulatory networks and diverse over molecule interactions with their context information. From the integrated data, we develop a generic virtual physiological human system. With the application of simulation and inference techniques to our constructed system, we study the efficacy, side effects, and mechanisms of natural compounds and combination drugs for the treatment of diseases.
Bio-Data Mining Sponsors: Bio-Synergy Research Project of the Ministry of Science and ICT
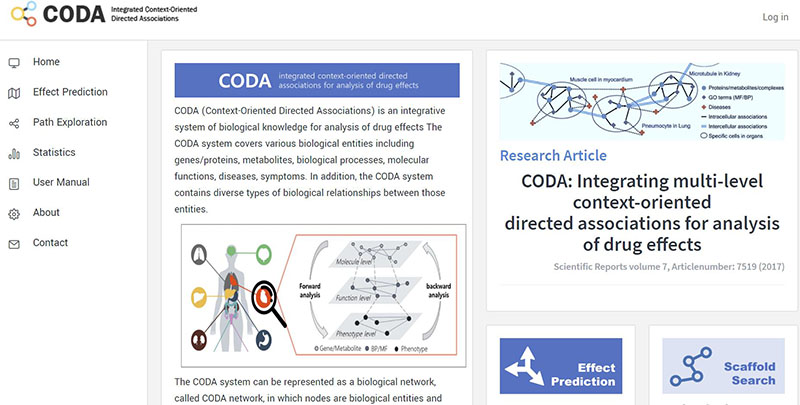
For most diseases, the single drug, single target model has limitation in many aspects. Furthermore, it is now evident that multi-component drugs or multi-targeting drugs, work synergistically to potentiate therapeutic effects of multiple targets and signaling pathways. Synergistic action of such drugs may overcome side effects that resulted from high doses of single-target drugs, increase drug selectivity, and offer an opportunity for more precise control of biological systems. In fact, in diverse therapies based on coordinated actions of multiple compounds have been invaluable sources of drug discovery and development. Thus, we designed and constructed the integrated database, which contains traditional herbal medicine, functional food, and up-to-date molecular drug/target information. Moreover we provide a search engine which makes it possible to infer compound combinations with clinical effects by applying data mining processes.
Bio-Visualization Sponsors: Bio-Synergy Research Project of the Ministry of Science and ICT
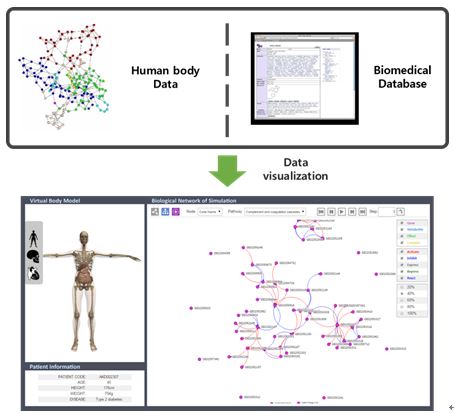
Biological system is very complex because various biomolecules and a lot of interactions are involved in the system. There are various types of biological interactions in our human body system. To understand whole biological systems properly, many types of biological interactions must be integrated and analyzed comprehensively. In this situation, we study method for the effective visualization of biological interaction data to handle and analyze the large biological systems easily.
- 1. “Meta-path Based Prioritization of Functional Drug Actions with Multi-Level Biological Networks,” Scientific Reports 2019
- 2. “Phenotype-oriented network analysis for discovering pharmacological effects of natural compounds,” Scientific Reports 2018
- 3. “CONET: a virtual human system-centered platform for drug discovery,” Frontiers in Computer Science 2018
- 4. “CODA: Integrating multi-level context-oriented directed associations for drug effects analysis,” Scientific Reports 2017
- 5. “Normalization of Tumor Vessels by Tie2 Activation and Ang2 Inhibition Enhances Drug Delivery and Produces a Favorable Tumor Microenvironment,” Cancer Cell 2016
- 6. “Inferring pathway activity toward precise disease classification,” PLoS Computational Biology 2008
- 7. “Network-based classification of breast cancer metastasis,” Molecular Systems Biology 2007