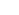"Synergistic Bioinformatics lab" aims to discover the best candidates of disease markers and therapeutic drugs integrating IT, BT, and computational chemistry. We construct artificial intelligence systems assimilating disease mechanisms which facilitate the prediction of disease marker and drug candidates. We also develop the cellular surrogate systems specific to the disease mechanisms which enable in vitro validation of the markers and drugs for the patients with the same mechanisms.
Cellular Regulatory Network informatics for inferring “Synergy-Markers”
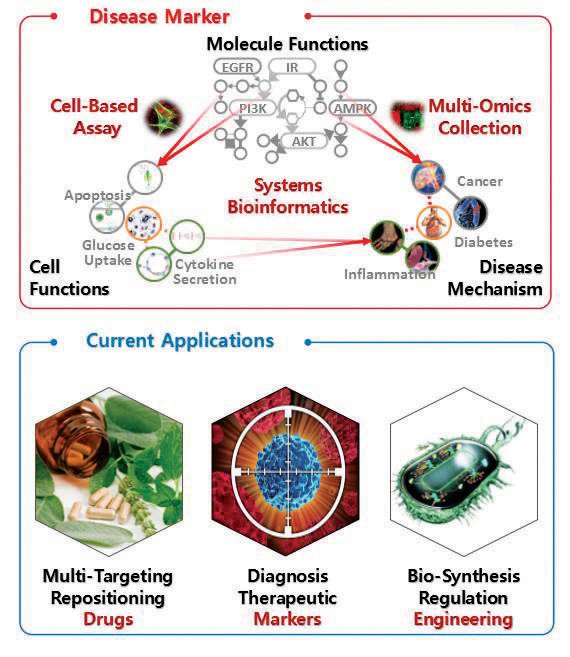
We build a cellular regulatory network with the relations among the genes from signaling and metabolic pathways including kinase/phosphatase-targets, E3 ligase-targets, TF-targets or epigenetic regulation targets, protein interactions, and scaffold-targets. We also build a functional module set with the gene set correlated together with any level of cellular functions in a cell from such as the gene sets in each GO term, co-expressed or co-identified in any specific cell function or subcellular location.
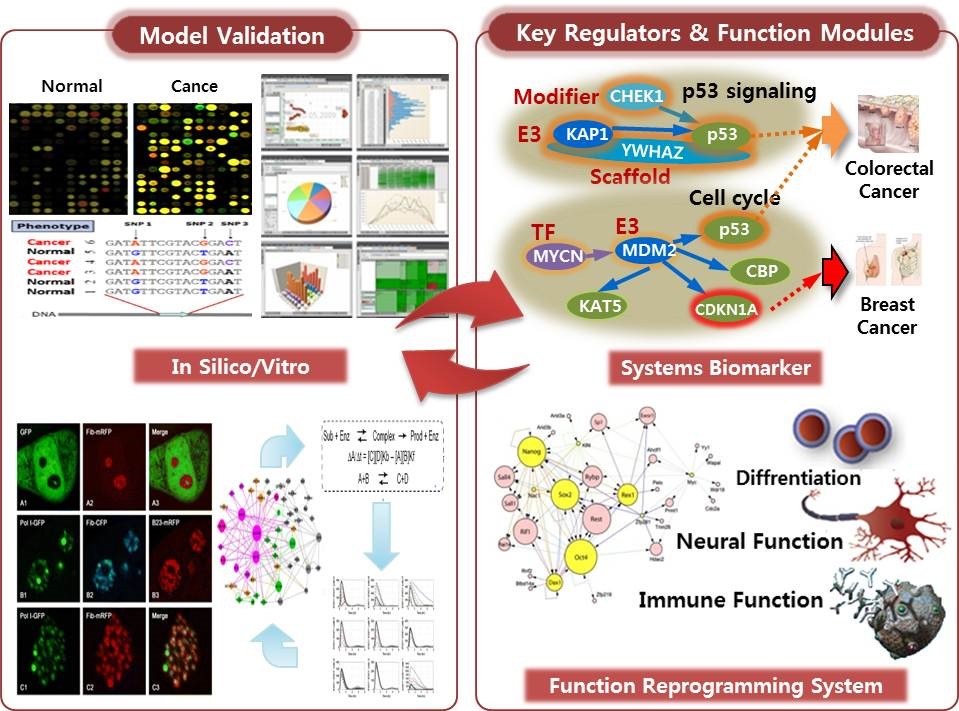
The information in the “Disease Associated Cellular Regulatory Network” is integrated to a systematic process to predict the significance of genes and their functional modules in specific disease models. Various information for disease association of genes and disease specific omics data sets are used to derive the best markers or therapeutic regulators, the “Synergy-Markers” that can represent or regulate the activities of mechanisms in specific disease models or patient groups.
Artificial Intelligence for inferring “Synergy-Drugs”
The process of “Synergy-Markers” is connected to identify the best therapeutic candidates, “Synergy-Drugs” from the known drugs or various chemical libraries. For this, we use various artificial intelligence methods to related the primary, secondary and tertiary structures of chemicals and biomolecules and their various levels of cellular mechanisms to their specific functional outputs. In this process, we infer the most possible responses of genes, functional modules, and mechanisms in the specific disease models targeted by individual or associated compounds or genes, or reversely the most possible compounds or genes promoting those responses
To maximize the performance, currently, we integrate several deep learning models as they assimilate the cellular regulatory models and also make up the lack of data sets. The semi-supervised learning model and auto- encoder model are applied to expand the learning data sets. Transfer learning model is applied to optimize the step-wise learning of each level off disease mechanism. The traceable or explainable learning formats are included to verify and enhance the benefits of assimilating underlying biological mechanisms. Capsule network model is one example that we can directly assimilate the hierarchical structure of cellular mechanisms on the learning model. Various methods to implement the cellular regulatory networks and pre-defined knowledge about drug or disease mechanisms are developed and tested. The success of these works can be a guide to solve some major huddles of artificial intelligence technology too.
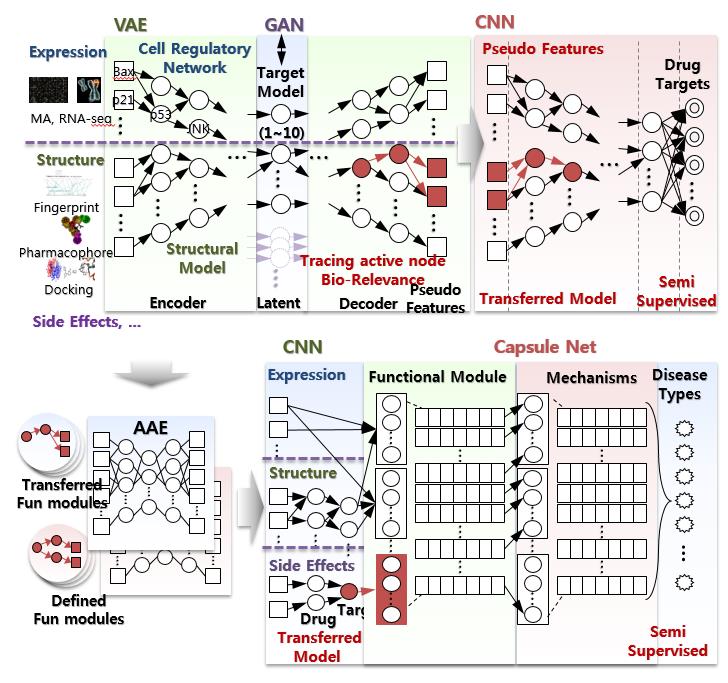
Cellular Surrogate System for Validation
We try to develop the best cellular surrogate system for the in vitro validation of disease models by analyzing the various realization of “Synergy-Markers” in the cell line models. The specific cellular mechanisms presented by each cell line are selected and grouped to represent the patient model. The changes of mechanism signatures by therapeutic agents in those cell lines are tested to predict the effect of those agents on patients. We are trying to find the benefit of this protocol in the drug repositioning, identifying the objects for companion diagnosis, or therapeutic target regulators for a specific cellular functions.
Engineering Prospective
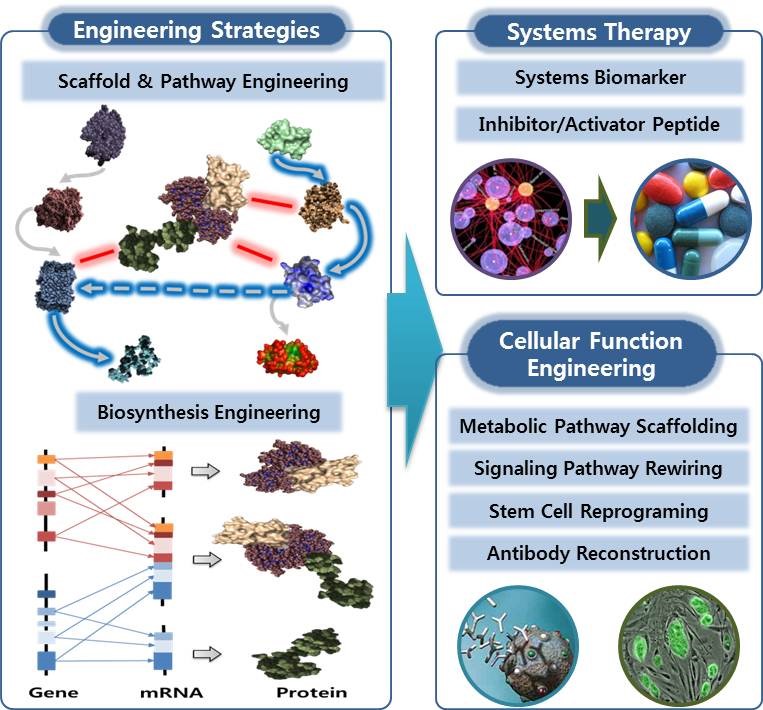
Our comprehensive and synergistic methods delineating the cellular mechanisms from the molecular structural details to the regulatory networks of cellular functions will support to construct more reasonable solutions for the current problems in biomedical fields that cannot be solved by individual method or skill sets. The validated compounds or regulator genes can be used to manipulate cellular functions. Those agents can be engineered for the better and more specific performance based on their possible implications in the structures, molecular relations, and functional mechanisms obtained from our processes.
Our primary engineering targets are the multi-functional and multi-targeting compounds or proteins that improve the detection or manipulation of the cellular functions, pathways or regulation circuitry with high fidelity and flexibility. The results will provide more refined solutions for the diagnosis or therapy and also in the other applications in biotechnology.