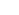Recent advances in high-throughput sequencing and chip-based technologies have brought a paradigm shift to the way we study underlying mechanisms of life. In the sea of ever growing genomics data, we can now ask and answer novel questions that have never been asked or answered before, thanks to the phenomenal development of computer and information technologies. The aim of our laboratory is to uncover novel knowledge that is buried under a wealth of biological information by applying computational or statistical methods to whole-genome genetic, epigenetic, and transcriptional data, hoping for valuable discoveries that can increase human health.
Research Keywords
Genomics, Epigenomics, Systems genetics, Machine learning
What is systems genetics?
The defining principle of systems genetics is understanding how genetic information is integrated, coordinated, and ultimately transmitted through molecular, cellular, and physiological networks to enable the higher-order functions and emergent properties of biological systems
Why omics?
Biology has become a data-rich subject, with genome-scale studies occurring in all fields. Ever since the rise of genomics, the suffix ‘-omics’ has been added to the names of many fields to denote studies undertaken on a large or genome-wide scale.
Omics is a general term for a broad discipline of science and engineering for analyzing the interactions of biological information objects in various ‘omes’.
The main focus is on:
1) mapping information objects such as genes, proteins, and ligands;
2) finding interaction relationships among the objects;
3) engineering the networks and objects to understand and manipulate the regulatory mechanisms;
4) integrating various omes and omics subfields.
- 1. Epigenetic regulation and the variability of gene expression (2008) Nature Genetics
- 2. Intrinsic variability of gene expression encoded in nucleosome positioning sequences (2009) Nature Genetics
- 3. Selected heterozygosity at cis-regulatory sequences increases the expression homogeneity of a cell population in humans (2016) Genome Biology
- 4. Histone variant H3F3A promotes lung cancer cell migration through intronic regulation (2016) Nature Communications
- 5. Chromatin structure-based prediction of recurrent noncoding mutations in cancer (2016) Nature Genetics