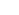“We decode bio-complexity to unravel/control the hidden logic of life and to further apply it for engineering innovation.” The Life Sciences are witnessing a shift of paradigm from traditional characterization of individual molecules towards an understanding of interactive pathways and networks. The role of genes, proteins, metabolites and cells can be understood and defined through their interactions and it is through our focus on intra- and inter-cellular dynamics that we are deeply involved in the emerging area of Systems Biology. Three key characteristics of Systems Biology are dynamic modeling, analysis, and control of complex biological networks. In this context, we focus on developing a systems biological analysis of biological information processing by signaling and gene regulation with particular emphasis towards understanding/controlling cell-fate determination. Our research is driven by three long term objectives: (1) understanding and controlling of complex biological phenomena; (2) developing practical biomedical technologies for personalized therapy and systems healthcare; (3) identifying novel drug targets for complex human disease. In this way, we are developing innovative theory and useful technologies in systems biology and bio-inspired engineering by combining biology and engineering.
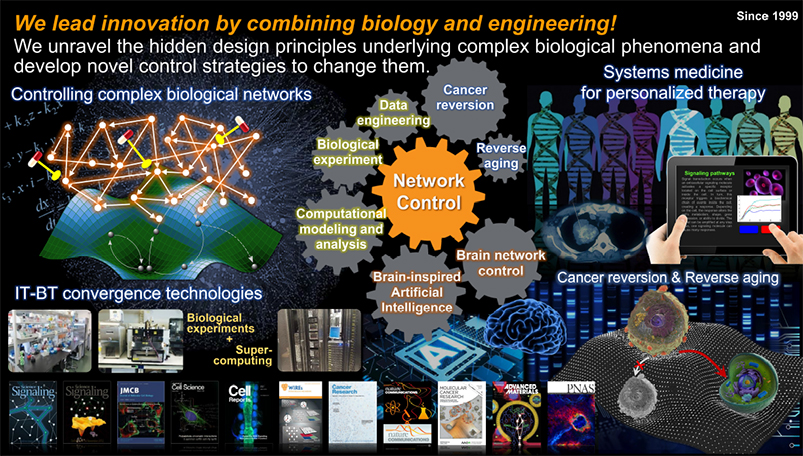
Reverse Control of Cancer and Aging
Cancer and aging are generally regarded as irreversible biological phenomena. Might it be possible to reverse these processes? Historically, there have been some reports about such reversion under particular circumstances, indicating the possibility of it, but no systematic analysis or experiment has been attempted so far. We are conducting very creative research on reverse control of cancer and aging through Systems Biology approach to challenge this problem.
Through extensive large-scale computer simulation analysis of these models, we try to identify molecular targets that can reverse cancer and aging processes and to further test the resulting changes in dynamic features when those targets are controlled. We also carry out experimental validation of the results through both cell and organoid experiments.
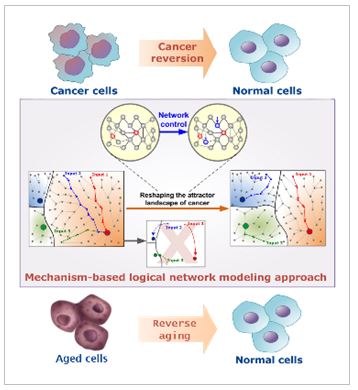
Computer Simulation Analysis for Personalized Therapy of Cancer
We are developing computer simulation models for primary molecular regulatory networks of cancer on the basis of biological big data. By mapping individual patient’s genetic variation to our simulation model, we can use our model to finding out optimal personalized therapy. Individually optimized therapeutic strategies are investigated by analyzing the dynamical features of our personalized computer simulation model through supercomputing-based large-scale simulation analysis. Our study suggests a new paradigm for precision medicine.
Analysis and Control of Biological Networks
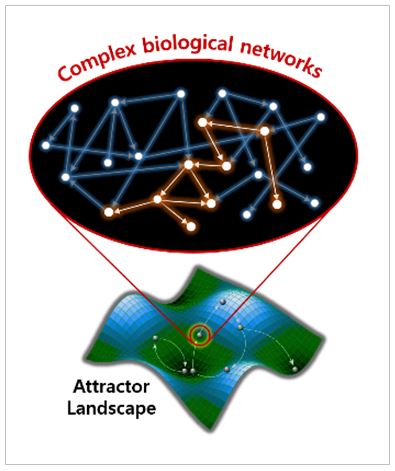
All living-systems are composed of complex networks of their functional subunits and their biological phenomena are determined by dynamic changes of those networks. Therefore, it is indispensable to control those complex networks to regulate the biological phenomena in the way we want. We explore the functions of such biological networks by analyzing both topological as well as dynamical properties. The aim of network systems biology is to investigate the relationship between topological properties and biological function, and to eventually develop practically useful control strategies with which we can regulate the network dynamics.
Brain Network Control
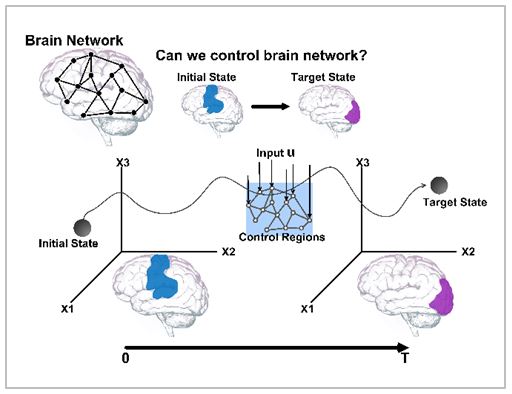
Our brain comprises complex networks of neurons through synaptic interactions and brain functions are induced by the dynamics of such complex networks. For instance, the selective synchronization is a basis for normal brain functioning, so synchrony disruption can cause functional abnormalities that lead to diverse brain disorder. We investigate the physical and functional regulations in brain and principles of those regulations using large-scale computational models of neuronal networks on the basis of multi-modal brain connectome data. The ultimate goal is to investigate the emergent properties of complex neuronal networks in connection with brain functions and disorders, and to unravel/control the underlying hidden working principles. For this purpose, we are constructing real human brain network models based on the human connectome data and developing brain network control strategies to overcome various brain disorders.
Bio-Inspired Engineering Based on Systems Biology
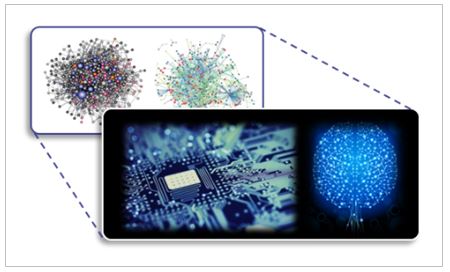
We are developing a new realm of engineering, named bio-inspired engineering based on Systems Biology, which applies our system-level understanding on biological mechanisms to engineering to find out a novel solution to unsolved engineering problems. For example, we develop self-repairing electrical circuits inspired from biological networks.
-
1. N. Kim, C. Y. Hwang, T. Kim, H. Kim, and K.-H. Cho*, "A cell fate reprogramming strategy reverses epithelial-to-mesenchymal transition of lung cancer cells while avoiding hybrid states", Cancer Research, Jan. 2023.
2. D. Shin and K.-H. Cho*, "Critical Transition and Reversion of Tumorigenesis", Experimental and Molecular Medicine, Jan. 2023.
3. K.-W. Lee, S.-Y. Yeo, J.-R. Gong, O.-J. Koo, I. Sohn, W. Y. Lee, H. C. Kim, S. H. Yun, Y. B. Cho, M.-A. Choi, S. An, J. Kim, C. O. Sung*, K.-H. Cho*, S.-H. Kim*, “PRRX1 is a master transcription factor of stromal fibroblasts for myofibroblastic lineage progression”, Nature Communications, 13, 2793, pp. 1-23, May, 2022.
4. S.R. Choi, C.Y. Hwang, J. Lee, and K.-H. Cho*, “Network analysis identifies regulators of basal-like breast cancer reprogramming and endocrine therapy vulnerability”, Cancer Research, 82, 2, pp. 320-330, Jan. 2022.
5. J. C. Park, S. Y. Jang, D. Lee, J. Lee, U. Kang, H. Chang, H. J. Kim, S. H. Han, J. Seo, M. Choi, D. Y. Lee, M. S. Byun, D. Yi, K.-H. Cho* and M. J. Inhee*, "A logical network-based drug-screening platform for Alzheimer’s disease representing pathological features of human brain organoids", Nature Communications, 12, 1, pp. 1-13, Jan. 2021.
6. S. An, S. Y. Cho, J. Kang, S. Lee, H. S. Kim, D. J. Min, E. Son, and K.-H. Cho*, "Inhibition of 3-phosphoinositide–dependent protein kinase 1 (PDK1) can revert cellular senescence in human dermal fibroblasts", Proceedings of the National Academy of Sciences, 117, 49, pp. 31535-31546, Nov. 2020.
7. M. P. Menden, et al. (K.-H. Cho, S. Cho, Y. Han, J. Kim, Y. Kim, J.-H. Song as members of AstraZeneca-Sanger Drug Combination DREAM Consortium), "Community assessment to advance computational prediction of cancer drug combinations in a pharmacogenomic screen", Nature Communications, 10, 1, pp. 1-17, Jun. 2019.
8. B. Lee, D. Shin, S. P. Gross, and K.-H. Cho*, "Combined positive and negative feedback allows modulation of neuronal oscillation frequency during sensory processing", Cell Reports, 25, 6, pp. 1548-1560, Nov. 2018.
9. S. Y. Yeo, K. W. Lee, D. Shin, S. An, K.-H. Cho* and S. H. Kim*, "A positive feedback loop bi-stably activates fibroblasts", Nature Communications, Vol. 9:3016, pp. 1-16, Aug. 2018.
10. M. Choi, J. Shi, Y. Zhu, R. Yang, and K.-H. Cho*, "Network dynamics-based stratification of cancer panel for systemic prediction of anticancer drug response", Nature Communications, 8, 1940, pp. 1-12, Dec. 2017.
11. D. Shin, J. Lee, J.-R. Gong, and K.-H. Cho*, "Percolation transition of cooperative mutational effects in colorectal tumorigenesis", Nature Communications, 8, 1270, pp.1-14, Nov. 2017.
12. J. K. Won, S. J. C. Y. Yu, Hwang, S. H. Cho, S. M. Park, K. Kim, W.-M. Choi, H. Cho, E. J. Cho, J.-H. Lee, K. B. Lee, Y. J. Kim, K.-S. Suh, J.-J. Jang, C. Y. Kim, J.-H. Yoon, and K.-H. Cho*, "Protein disulfide isomerase inhibition synergistically enhances the efficacy of sorafenib for hepatocellular carcinoma", Hepatology, 66, 3, pp. 855-868, Jul. 2017.
13. K.-H. Cho*, S. Lee, D. Kim, D. Shin, J. I. Joo, and S.-M. Park, "Cancer reversion, a renewed challenge in systems biology", Current Opinion in Systems Biology, 2, pp. 48-57, Apr. 2017.
14. K.-H. Cho*, J. I. Joo, D. Shin, D. Kim, and S.-M. Park, "The reverse control of irreversible biological processes", WIREs Systems Biology and Medicine, 8, 5, pp. 366-377, Sep/Oct. 2016.
15. S.-Y. Shin, T. Kim, H.-S. Lee, J. H. Kang, J. Y. Lee, K.-H. Cho* and D. H. Kim, "The switching role of β-adrenergic receptor signaling in cell survival or death decision of cardiomyocytes", Nature Communications, Vol. 5:5777, pp. 1-13, Dec. 2014.
16. H.-S. Lee, C. Y. Hwang, S.-Y. Shin, K.-S. Kwon, and K.-H. Cho*, "MLK3 is part of a feedback mechanism that regulates different cellular responses to reactive oxygen species", Science Signaling, Vol. 7, Issue 328, ra52, Jun. 2014.
17. M. Choi, J. Shi, S. H. Jung, X. Chen, and K.-H. Cho*, "Attractor Landscape Analysis Reveals Feedback Loops in the p53 Network That Control the Cellular Response to DNA Damage", Science Signaling, Vol. 5, Issue 251, ra83, Nov. 2012.
18. J.-R. Kim, J. Kim, Y.-K. Kwon, H.-Y. Lee, P. Heslop-Harrison, and K.-H. Cho*, "Reduction of Complex Signaling Networks to a Representative Kernel", Science Signaling, Vol. 4, Issue 175, ra35, 31-May 2011.
19. S.-Y. Shin, O. Rath, A. Zebisch, S.-M. Choo, W. Kolch, and K.-H. Cho*, "Functional roles of multiple feedback loops in ERK and Wnt signaling pathways that regulate epithelial-mesenchymal transition", Cancer Research, Vol. 70, Issue 17, pp. 6715-6724, Sept. 2010.